Conserved Drought Responses and Gene Expression in Sorghum and Maize
Drought stress costs billions of dollars in crop loss annually. Tolerance to drought is a complex trait which makes it difficult to identify the underlying genetic loci and the associated drought-responsive pathways. Core and deeply conserved drought responses not only operate in the sorghum germplasm, but also across related species such as Zea mays (maize) due to ancestral adaptations. To compare drought response and gene expression datasets between sorghum and maize as well as within each species, researchers from Michigan State University, Cornell University and the USDA-ARS developed a predictive model approach combined with differential expression analysis across 25 diverse sorghum genotypes and 27 diverse maize genotypes to study the ancestral adaptations of drought stress in C4 grasses. These studies found that core drought responsive genes are conserved between maize and sorghum and top predictors had a significantly higher proportion of genes with an average SIFT score above 0.05 than random background genes suggesting conserved genes are less likely to contain deleterious mutations. These genes were found enriched with GO terms of response to abiotic stimulus, reactive oxygen species scavenging enzymes as well as folding and refolding of proteins.
This study demonstrates a broad evolutionary conservation of drought responses across C4 grasses which could have important implications for developing climate resilient cereals.
Drought tolerance is a complex phenotype that is difficult to distill down into single causal genes.
We were very fortunate to capture a mild drought event in summer 2020 in Michigan. Seedlings from the sorghum association panel were clearly drought stressed from a physiological standpoint, but when we looked at gene expression, we saw no common genes across accessions. This led us to use a predictive modeling approach to see if we could identify emergent features underlying drought responses. To our surprise this worked quite well, and our model could even be applied to maize data.
We see our study as an exciting proof of concept that machine learning and predictive modeling can be used to identify important features underlying stress responses. In future work, we hope to use these approaches to identify physiological and gene expression signatures underlying climate resilience. – VanBuren
SorghumBase Examples:
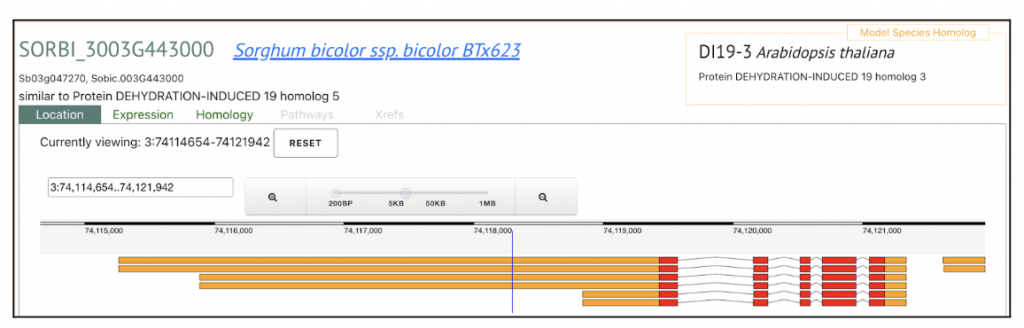

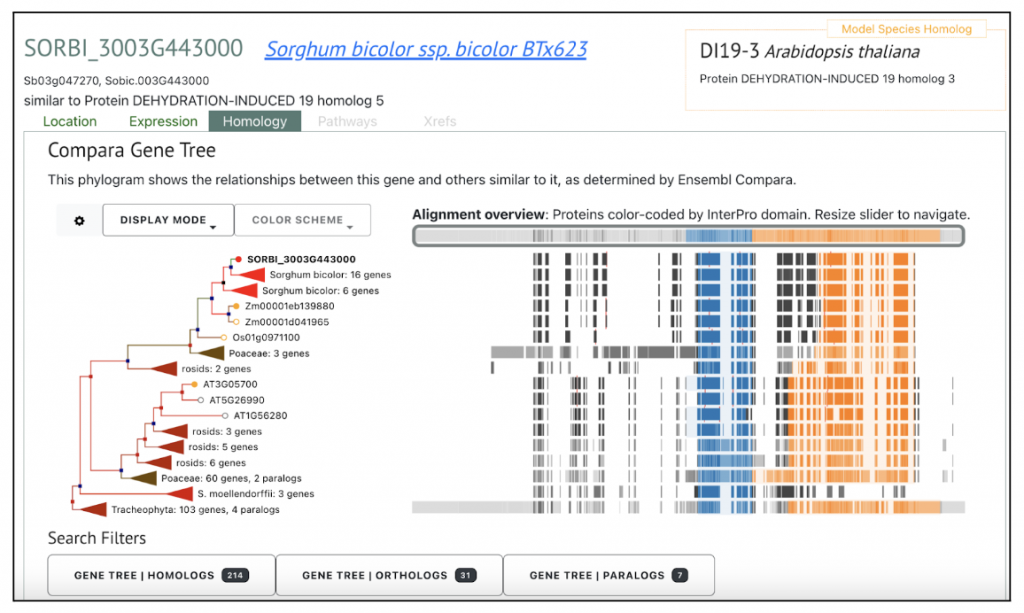
Reference:
Pardo J, Wai CM, Harman M, Nguyen A, Kremling KA, Romay MC, Lepak N, Bauerle TL, Buckler ES, Thompson AM, VanBuren R. Cross-species predictive modeling reveals conserved drought responses between maize and sorghum. Proc Natl Acad Sci U S A. 2023 Mar 7;120(10):e2216894120. PMID: 36848555. DOI: 10.1073/pnas.2216894120. Read more
Related Project Websites:
VanBuren Lab at Michigan State University: https://www.vanburenlab.org/
Thompson Lab at Michigan State University: www.thompsonmaizelab.org


