Integrative investigation shows promise for narrowing candidate genes in forage sorghum yield and other complex traits
Sorghum is a highly resilient crop with the ability to grow in adverse conditions and withstand salinity, drought, and waterlogging. It is useful for cultivation as a forage crop for livestock, especially because the domestic animal industry is growing and solutions are needed for feed in dry or semi-dry climates. The increase of yield in the sorghum crop is of special interest to breeders and owners of livestock.
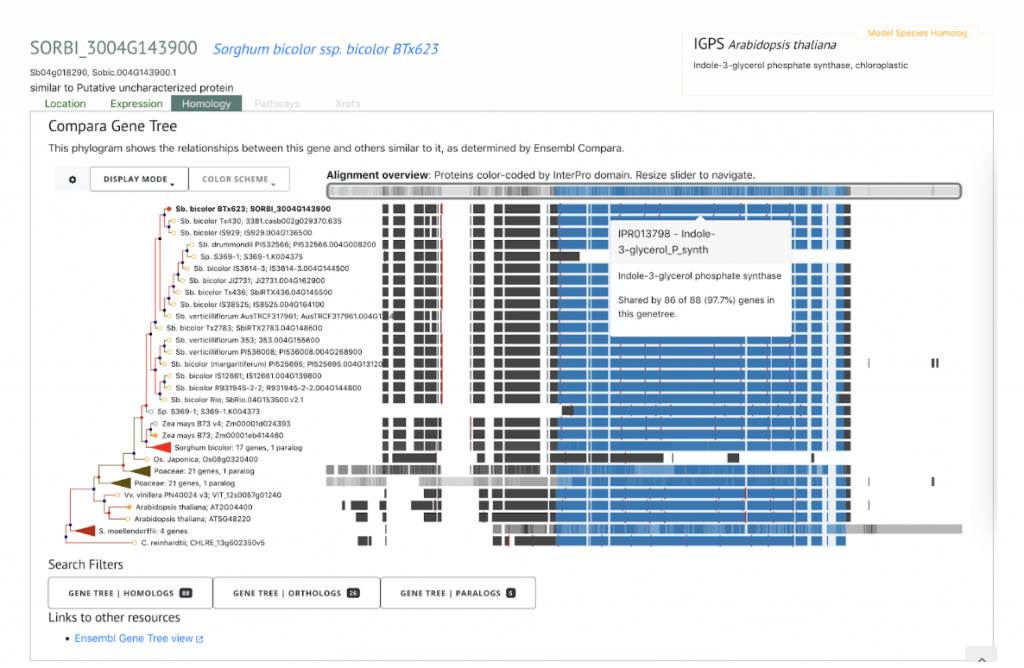
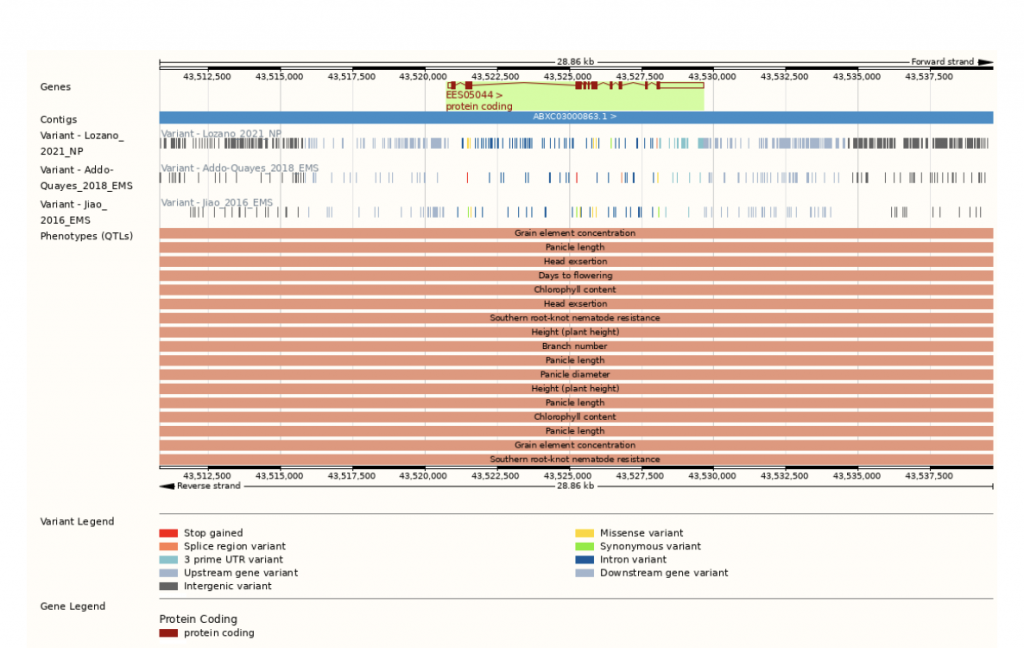
In an effort to increase sorghum’s forage yield, researchers from Nanjing Agricultural University in Nanjing, Anhui Science and Technology University in Fengyang, China and University of Louisiana at Lafayette collaborated to genetically analyze sorghum yield traits. Genome-wide association studies (GWAS) are frequently used to map complex traits, such as forage yield. In this study, over 85.5K SNPs were used to map forage yield, and RNA sequencing (RNA-Seq), an approach to quantify gene expression, was used to refine GWAS results and hone in on a shorter list of potential gene candidates at specific QTLs. Over the course of two years, plant height (PH), tiller number (TN), stem diameter (SD), and fresh weight per plant (FW) were analyzed on 245 sorghum accessions grown across four environments and two locations. 338 SNPs or more accurately quantitative trait nucleotides (QTNs) were identified, but only 21 occurred in at least two environments, indicating that performing the analysis on sorghum in multiple testing environments helps significantly narrow down the number of potential QTNs. When the results were compared with those from previous studies, 12 QTNs overlapped with the known QTLs (two for PH, six for TN and four for SD), providing strong, likely targets for gene cloning and breeding. There was a strong correlation between plant height and forage yield. Eight candidate genes were identified through this approach, including Sobic.004G143900, a gene involved in indoleacetic acid biosynthesis, that reaches its highest expression rate in young steam and is down-regulated as the plant matures. In addition, the study identified a potential association between the RAD52-like DNA repair gene Sobic.003G375100 and SD. Overall, this study demonstrates that the integrative approach to analyzing omics data from GWAS and RNA-Seq shows potential in the identification of candidate genes for complex traits and future transgenic studies.
SorghumBase examples:
Reference
Wang L, Liu Y, Gao L, Yang X, Zhang X, Xie S, Chen M, Wang YH, Li J, Shen Y. Identification of Candidate Forage Yield Genes in Sorghum (Sorghum bicolor L.) Using Integrated Genome-Wide Association Studies and RNA-Seq. Front Plant Sci. 2022 Jan 11;12:788433. PMID: 35087554. DOI: 10.3389/fpls.2021.788433. Read more

