Physiological and Transcriptomic changes under Salt Stress in ‘Sodamchal’, a Korean Line of Sorghum
Salt stress impacts 20% of cultivable land as well as 50% of irrigated land. Exposure to salt affects lipid metabolism, photosynthesis, ion homeostasis, and hormones in plants, causing issues with growth and development, decreased yield and, at times, death. The prevalence of high soil salinity and its effect on yield make it a topic of great importance for scientists and growers.
In an effort to investigate the way genes are expressed under salt stress in sorghum, researchers from Chungnam National University, and the National Institute of Agricultural Sciences in Korea studied physiological and transcriptomic changes in ‘Sodamchal’, a line of Korean sorghum known for its salt sensitivity. The study subjected the plants in the experimental group to 150 mM of salt and compared the results with a control group exposed to 0 nM of salt. Leaves and roots were collected at 0, 3, and 9 days after the treatment. The physiological changes were investigated over time to determine if they would be a good indicator of the level of stress. Both anthocyanin and chlorophyll were determined to likely not be good candidates for stress indicators. Anthocyanin in both the control and salt exposure group increased between 0 and 9 days with peak concentrations at the 3 day point. Since both groups showed a similar pattern of significant changes, factors other than salt likely influenced the expression. Chlorophyll levels also showed similar patterns over time for the control and salt treatment groups and the fluctuations in the stressed group were not statistically significant. Reducing sugar and proline showed more promise. Reduction of sugar decreased over time in the salt stress group and is a likely candidate for evaluating salt stress on plants. While the control group did not show any significant changes in proline levels, the experimental group showed a large increase in proline over time. The homeostasis and osmotic pressure in cells is compromised with significant salt exposure and proline helps maintain osmotic pressure.The results indicate that proline may be the most optimal indicator of salt stress of the physiological factors investigated in this study.
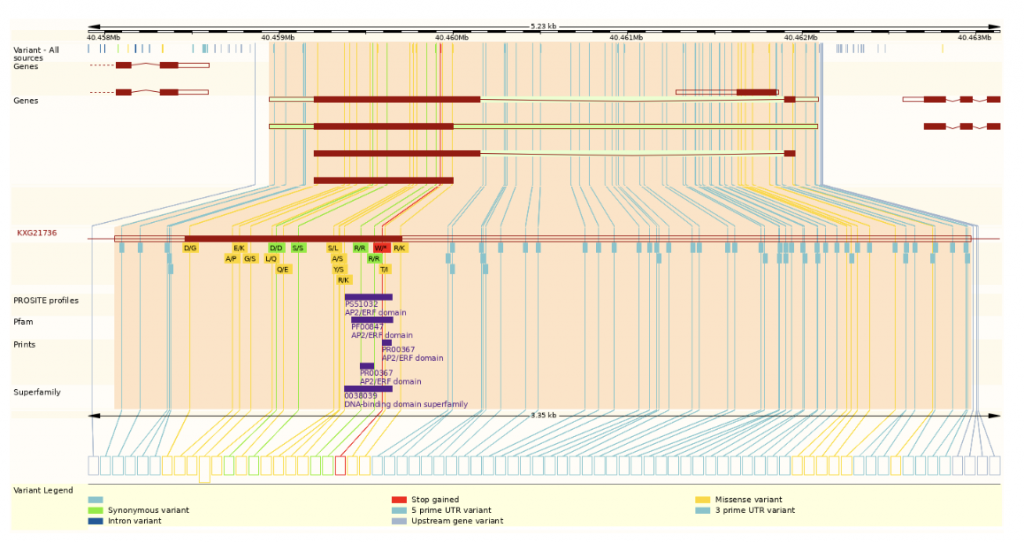
The transcription profile was evaluated using QuantSeq for both the control and the 150 mM salt condition to determine candidate genes for the salt stress group. There were 1506 differentially expressed genes (DEGs) between the two groups in the leaves and 1510 in roots. The gene co-expression network was used to identify up-regulated transcription factors AP2/ERF and Dehydrin as being associated with salt stress.
SorghumBase example
The authors confirmed that AP2/ERF (APETAL2/ethylene response factor) and Dehydrin (DHN) are transcription factors up-regulated in leaves and roots subject to high salinity conditions, abiotic stress.
Reference
Choi S, Kang Y, Lee S, Jeon DH, Seo S, Lee TH, Kim C. Physio-chemical and co-expression network analysis associated with salt stress in sorghum. Front Biosci (Landmark Ed). 2022 Feb 11;27(2):55. PMID: 35226998. DOI: 10.31083/j.fbl2702055. Read more
Related Project Websites:
https://biography.omicsonline.org/korea/chungnam-national-university/changsoo-lee-719179

