Sequenced Sorghum Mutant Library Investigated for Protein and Amino Acid Content Variation
Although sorghum is the fifth major cereal crop globally and is resistant to drought, its nutritional value is reduced when compared with wheat, maize and rice. This nutritional value is directly related to the grain’s protein quality. There is variation among the different lines, but on average sorghum contains 11% crude protein, ~70% starch and 2–4% lipids and other oils. Overall, sorghum has lower protein digestibility as well as low essential amino acids content when compared with these other cereals. Insufficient amino acid intake, which is common in people in countries with a high reliance on subsistence farming who depend on a limited number of crops for food, causes health issues. Adding chemically generated amino acids is not “cost effective,” making it crucial to increase the amino acid and protein content in the crops themselves. Although both mutant populations and wild-type lines are commonly used as sources for genetic diversity, ethyl methanesulfonate (EMS) mutant populations in sorghum have not been screened extensively for seed quality traits. Sorghum researchers from Texas Tech University and the USDA-ARS studied total protein content and 23 amino acids in 206 EMS-based sorghum mutant lines in an effort to provide a new germplasm resource for sorghum grain quality improvement. Their results showed variable protein and amino acid content in the EMS lines with a range of 10–19.2% for total protein content. Compared to the wild type BTx623, some of the EMS mutants had nearly double the total protein content. The total protein content was also positively correlated with the essential amino acid content. The mutant population can help pinpoint the genes responsible for seed quality. Those with significant changes in total protein and essential amino acids have the most potential to aid in this endeavor. The mutant lines that were produced will be freely available for use in breeding programs to improve seed quality in sorghum.
SorghumBase examples:
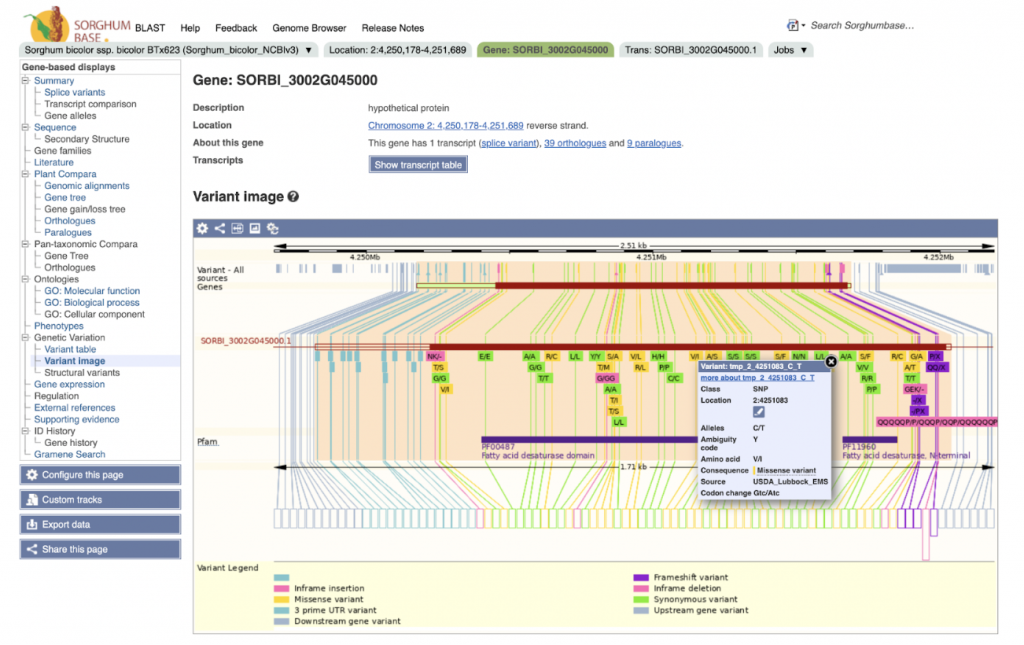
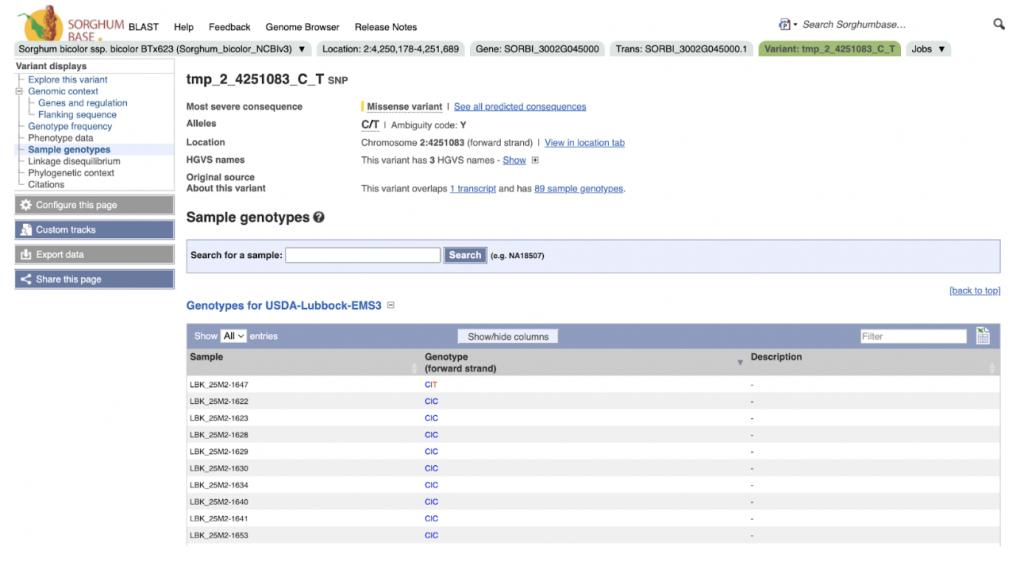
Reference:
Khan A, Khan NA, Bean SR, Chen J, Xin Z, Jiao Y. Variations in Total Protein and Amino Acids in the Sequenced Sorghum Mutant Library. Plants (Basel). 2023 Apr 15;12(8):1662. PMID: 37111885. DOI: 10.3390/plants12081662. Read more
Related Project Website:
Jiao Lab at Texas Tech University: https://www.depts.ttu.edu/igcast/Staff/jiao_lab.php


