Unique Chromatin Regulators Discovered in Sorghum
Sorghum, an important cereal crop that likely originated in Africa, has characteristic features and is highly stress tolerant. Epigenetic regulation involves changes in genome accessibility which impact gene expression and is believed to be behind the shaping of many of these unique features. Growers, breeders and scientists are greatly interested in understanding epigenetic regulation of gene expression and how it results in features that allow sorghum to thrive in stressed environments.
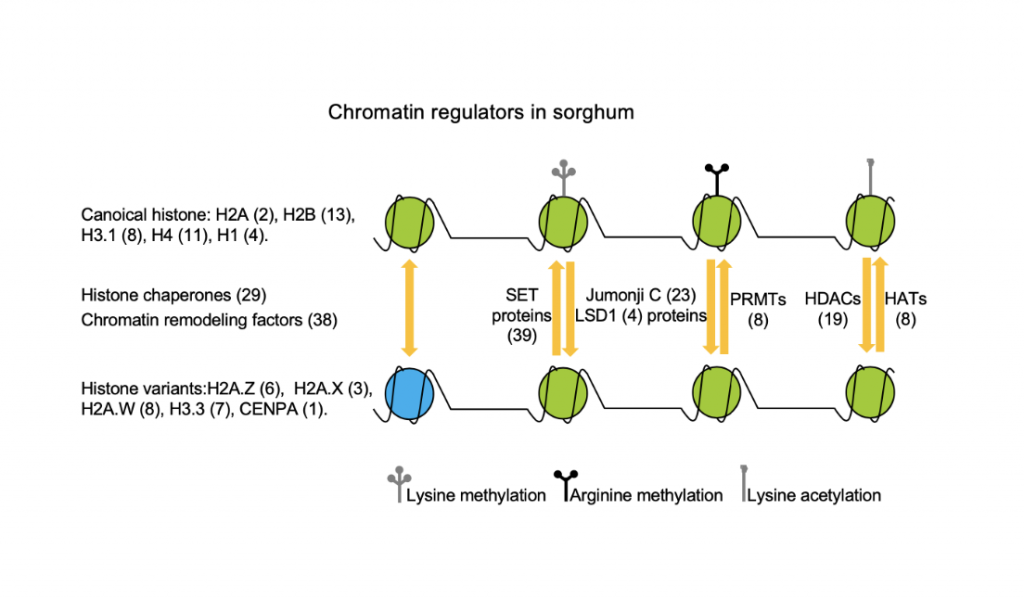
In an effort to prove that epigenetics is behind some of sorghum specific features, researchers at Three Gorges University in China sought to clarify the function of the chromatin regulators and their interaction network. Chromatin regulators influence the structure of chromatin, which has a critical effect on transcription, DNA replication and repair, and recombination. The researchers identified sorghum major chromatin regulators including 231 chromatin regulators of which there were 63 histones, 29 histone chaperones, 101 histone modification enzymes, and 38 chromatin remodeling factors. They found that the sorghum genome contains most of the genes for chromatin regulators found in Arabidopsis and rice, the result of differential duplication events of these common genes. There were however some novel histone proteins that evolved in sorghum and some other species of grass. They discovered a protein unique to sorghum that shares the domains of Snf2 family proteins, the elongation factor EF-Tu and the histone chaperone SPT16. It is unknown at this time whether any of these proteins are linked to phenomic characteristics, but this would be a good focus of future study. The researchers additionally separated the expression patterns of all the chromatin regulators into four categories in order to attempt to predict their functional interaction. More work is needed to completely understand how epigenetic regulation is influenced by the interaction between the chromatin regulators and how this results in the development and stress response in sorghum.
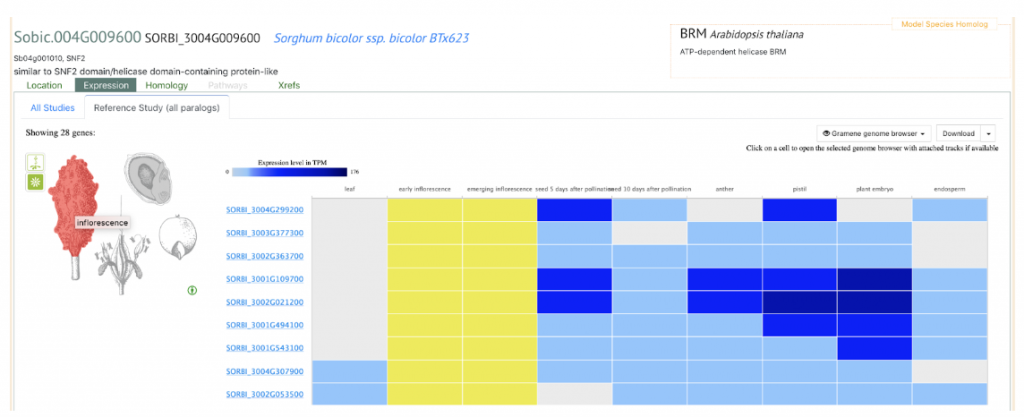
SorghumBase example:
Reference
Hu Y, Chen X, Zhou C, He Z, Shen X. Genome-wide identification of chromatin regulators in Sorghum bicolor. 3 Biotech. 2022 May;12(5):117. PMID: 35547013. DOI: 10.1007/s13205-022-03181-8. Read more