Assembly and Annotation of CP-NAM Parental Genotypes to Support Bioenergy Sorghum Research
The genetic diversity, adaptability and drought tolerance of sorghum (Sorghum bicolor (L.) Moench) make it of great use as a sustainable, fast-growing, and high-yielding bioenergy crop. Selectively breeding sorghum for further improvement requires knowledge of its genetic underpinnings, especially in terms of yield, carbon partitioning, and local adaptation. The complex nature of the genetic contributions to these traits makes it difficult to fully understand them. In fact, these kinds of traits often consist of both, changes in large numbers of genes, and complex structural mutations, in addition to single nucleotide polymorphisms (SNPs). To expand on a recently developed mapping population, scientists from Clemson University, Carolina Seed Systems, the University of North Carolina at Charlotte, Cold Spring Harbor Laboratory, and USDA-ARS, assembled and annotated genomes of the ten parental lines of the CP-NAM population. Over 24 thousand large structural variants (SVs) and over 10.5 million SNPs were identified. In particular, SVs and SNPs associated with phenotypes advantageous to sorghum’s use in bioenergy were focused on, as well as the variation between the sweet and cellulosic genotypes. The researchers found that SVs within coding regions impacted different types of genes compared to SNPs indicating that looking at both is of great importance when mapping traits. These discoveries will contribute to future mapping and trait discovery for sorghum. This new, extensive dataset is available through SorghumBase.
Our work here shows that long read sequencing and high-quality genome assemblies are vital to the understanding and improvement of sorghum as a potential source of sustainable biofuel in the future. We believe that there is a great necessity to explore genome-wide patterns of variation among crop species to determine the genetic architecture of complex traits, and we hope that this dataset and analysis can help contribute to future breeding efforts in sorghum. – Cooper and Voelker
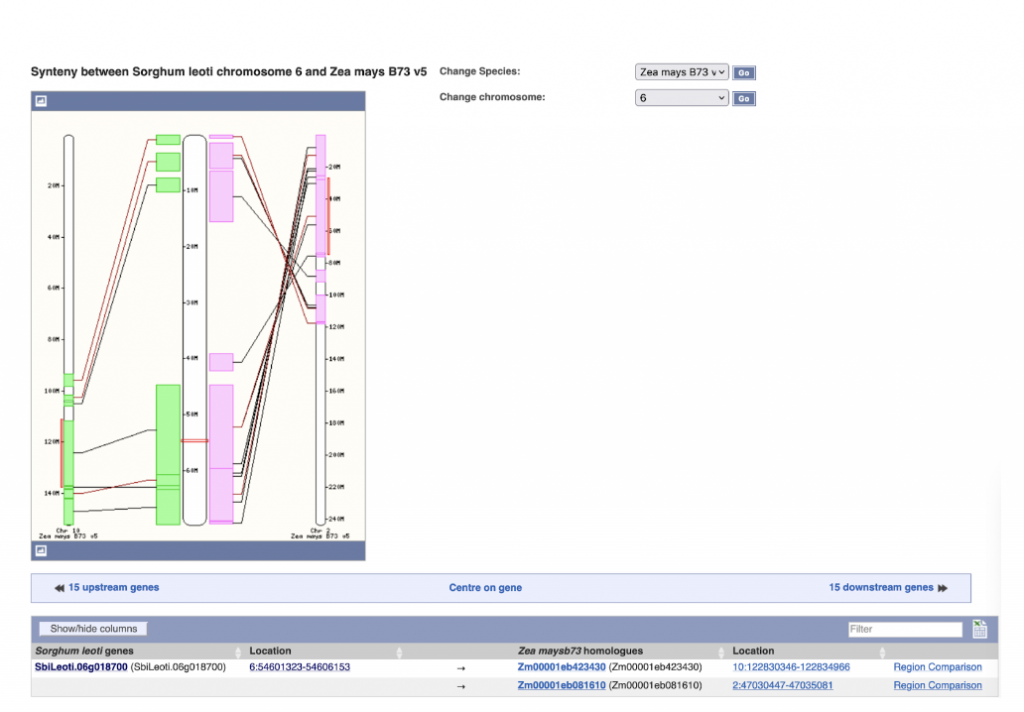
SorghumBase Examples:

Reference:
Voelker WG, Krishnan K, Chougule K, Alexander LC Jr, Lu Z, Olson A, Ware D, Songsomboon K, Ponce C, Brenton ZW, Boatwright JL, Cooper EA. Ten new high-quality genome assemblies for diverse bioenergy sorghum genotypes. Front Plant Sci. 2023 Jan 4;13:1040909. PMID: 36684744. DOI: 10.3389/fpls.2022.1040909. Read more
Related Project Websites:
Cooper Lab at UNC Charlotte: https://elizcooperlab.com/



